#include <string>#include "colarray/matrix.h"#include "colarray/vector.h"#include "tetmesh.h"#include "femaxmesh.h"#include "nedelecmesh.h"Include dependency graph for h5_tools.h:

This graph shows which files directly or indirectly include this file:
Go to the source code of this file.
Namespaces | |
| namespace | Teuchos |
Functions | |
| void | h5_create_empty_file (const std::string &file_name) |
| void | h5_write_param_list (const std::string &file_name, const Teuchos::ParameterList ¶ms) |
| void | h5_read_param_list (const std::string &file_name, Teuchos::ParameterList ¶ms) |
| void | h5_write_eigenmodes (const std::string &file_name, const Epetra_MultiVector &Q, const Epetra_SerialDenseVector &lambda) |
| void | h5_read_eigenmodes (const std::string &file_name, colarray::Matrix< double > &Q, colarray::ColumnVector< double > &lambda) |
| void | h5_read_eigenmesh (const std::string &file_name) |
| void | h5_write_eigenmesh (const std::string &file_name, const Epetra_Comm &comm, mesh::TetMesh *tetmesh) |
| void | h5_write_eigenpoint (const std::string &file_name, const Epetra_Comm &comm, mesh::TetMesh *tetmesh) |
| Write the tetrahedral point coordinates in parallel mode, ensuring unique file access and semantics. | |
| void | h5_write_eigenfield (const std::string &file_name, const Epetra_Comm &comm, mesh::TetMesh *tetmesh, NedelecMesh &nedelecmesh, const Epetra_MultiVector &Q, double *lambda) |
| Write the sampled electric and magnetic fields into the HDF5 file; the function must be called collectively. | |
| void | h5_write_eigenvalue (const std::string &file_name, const Epetra_Comm &comm, mesh::TetMesh *tetmesh, NedelecMesh &nedelecmesh, const Epetra_MultiVector &Q, double *lambda) |
| Write the eigenvalues stored in 'lambda' into the HDF5 file. | |
| void | h5_cartesian_sampling (const std::string &file_name, const Epetra_Comm &comm, mesh::TetMesh *tetmesh, NedelecMesh &nedelecmesh, const Epetra_MultiVector &Q, double *lambda, double xmin, double xmax, const int nx, double ymin, double ymax, const int ny, double zmin, double zmax, const int nz) |
| evaluate the eigenmodal solution on a cartesian grid | |
| void | h5_cartesian_sampling_write_hdf5 (const Epetra_Comm &comm, const std::string &file_name, unsigned int nmode, std::vector< std::vector< std::vector< unsigned int > > > &exists, std::vector< std::vector< std::vector< std::vector< mesh::Vector3 > > > > &efield, std::vector< std::vector< std::vector< std::vector< mesh::Vector3 > > > > &hfield, double xmin, double xmax, const int nx, double ymin, double ymax, const int ny, double zmin, double zmax, const int nz) |
| processorwise store the eigenmodal solution, sampled on a cartesian grid, into the hdf5 file; nota bene: because the sequence of access to the hdf5 file is in no way ordered, we must make sure that spatial positions marked as existent by one processor are not overwritten as non-existent by another processor. This is ensured as follows: (1) the root process only initializes the complete 3-dimensional boolean array as false (2) all processors only write into the file if the respective boolean flag signals that the position exists on the local processor; otherwise they do nothing to the hdf5 file. | |
| void h5_cartesian_sampling | ( | const std::string & | file_name, | |
| const Epetra_Comm & | comm, | |||
| mesh::TetMesh * | tetmesh, | |||
| NedelecMesh & | nedelecmesh, | |||
| const Epetra_MultiVector & | Q, | |||
| double * | lambda, | |||
| double | xmin, | |||
| double | xmax, | |||
| const int | nx, | |||
| double | ymin, | |||
| double | ymax, | |||
| const int | ny, | |||
| double | zmin, | |||
| double | zmax, | |||
| const int | nz | |||
| ) |
evaluate the eigenmodal solution on a cartesian grid
*
| void h5_cartesian_sampling_write_hdf5 | ( | const Epetra_Comm & | comm, | |
| const std::string & | file_name, | |||
| unsigned int | nmode, | |||
| std::vector< std::vector< std::vector< unsigned int > > > & | exists, | |||
| std::vector< std::vector< std::vector< std::vector< mesh::Vector3 > > > > & | efield, | |||
| std::vector< std::vector< std::vector< std::vector< mesh::Vector3 > > > > & | hfield, | |||
| double | xmin, | |||
| double | xmax, | |||
| const int | nx, | |||
| double | ymin, | |||
| double | ymax, | |||
| const int | ny, | |||
| double | zmin, | |||
| double | zmax, | |||
| const int | nz | |||
| ) |
processorwise store the eigenmodal solution, sampled on a cartesian grid, into the hdf5 file; nota bene: because the sequence of access to the hdf5 file is in no way ordered, we must make sure that spatial positions marked as existent by one processor are not overwritten as non-existent by another processor. This is ensured as follows: (1) the root process only initializes the complete 3-dimensional boolean array as false (2) all processors only write into the file if the respective boolean flag signals that the position exists on the local processor; otherwise they do nothing to the hdf5 file.
| void h5_create_empty_file | ( | const std::string & | file_name | ) |
End of forward declarations
Definition at line 130 of file h5_tools.cpp.
Referenced by FemaxxDriver::calculate_eigenfields(), and FemaxxDriver::run().
| void h5_read_eigenmesh | ( | const std::string & | file_name | ) |
Read the mesh used for the eigenvalue calculations into HDF5 file. The function must be called by alle processes
Definition at line 449 of file h5_tools.cpp.
| void h5_read_eigenmodes | ( | const std::string & | file_name, | |
| colarray::Matrix< double > & | Q, | |||
| colarray::ColumnVector< double > & | lambda | |||
| ) |
Definition at line 404 of file h5_tools.cpp.
References colarray::Matrix< T >::_v, lambda, num_eigenpairs(), and Q.
Here is the call graph for this function:
| void h5_read_param_list | ( | const std::string & | file_name, | |
| Teuchos::ParameterList & | params | |||
| ) |
Read parameter list to HDF5 file.
Parameters for the HDF5 are inserted into params.
Definition at line 280 of file h5_tools.cpp.
Referenced by main().
| void h5_write_eigenfield | ( | const std::string & | file_name, | |
| const Epetra_Comm & | comm, | |||
| mesh::TetMesh * | tetmesh, | |||
| NedelecMesh & | nedelecmesh, | |||
| const Epetra_MultiVector & | Q, | |||
| double * | lambda | |||
| ) |
Write the sampled electric and magnetic fields into the HDF5 file; the function must be called collectively.
To evaluate the electric and magnetic field at any location within the computational domain, we need the degrees of freedom (DoF) associated with the respective elements; the DoF are the elements of the eigenvectors, resulting from the numerical solution; this eigenvector is an Epetra_MultiVecor, distributed over the processes; therefore, we must make sure that we have on a specific process all the required elements of the eigenvector, especially, we must retrieve the DoF belonging to tetrahedra with facets and edges on interprocessor boundaries; this is accomplished by importing these eigenvector elements with the Epetra import and export commands, cf. the Trilinos tutorial for an introduction. 2006 feb 02, oswald.
store the rank of this prcocess within this MPI communicator
store the size of an MPI communicator, i.e. the number of processes engaged
Retrieve access to paralleltetmesh class instance
Retrieve fine element basis function for the edges and faces, via NedelecMesh.
nedelecmesh is a singleton
Open file collectively and release property list identifier
Retrieve FE approximation order and number of DoF
Retrieve finite element approximation order, valid throughout the mesh
Retrieve number of DoF associated with the element
Build a new vector to hold required elements
Build an import object for getting the required distributed vector elements from the other processes
Import the distributed vector elements
Retrieve local number of tetrahedra
Retrieve location at which electric field shall be sampled
allocate memory to store sampled electric field vectors
allocate memory to store sampled magnetic field vectors
Allocate 1D array for storing the electric field
Allocate 1D array for storing the magnetic field
Used as index for tetrahedra
Retrieve location at which electric field shall be sampled
Retrieve DoF vector required for base function evaluation; based on code in nedelecmesh::eval(...)
retrieve a pointer to the Tet class instance, behave, tit is an iterator, not a pointer
Evaluate the electric field
Evaluate the magnetic field
Store the electric field into an HDF5 file using parallel access mode; the algorithm is similar to the one used in the h5_write_eigenmesh function; we loop over the tetrahedra and store the corresponding electric field vector into the HDF5 file.
Loop over tetrahedra and store electric field into HDF5 file
Free dynamically allocated variables, if any
Definition at line 797 of file h5_tools.cpp.
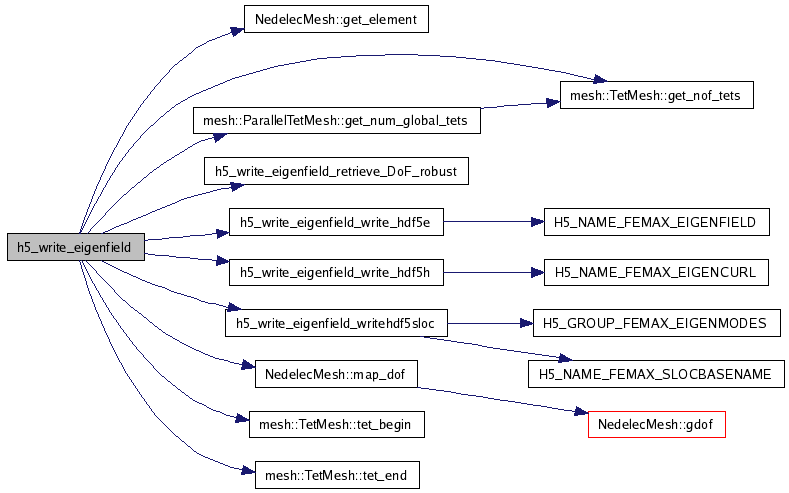
References NedelecMesh::get_element(), mesh::TetMesh::get_nof_tets(), mesh::ParallelTetMesh::get_num_global_tets(), h5_write_eigenfield_retrieve_DoF_robust(), h5_write_eigenfield_write_hdf5e(), h5_write_eigenfield_write_hdf5h(), h5_write_eigenfield_writehdf5sloc(), mesh::ID_NONE, NedelecMesh::map_dof(), NREALCOORD3D, mesh::TetMesh::tet_begin(), mesh::TetMesh::tet_end(), x, mesh::Vector3::x, mesh::Vector3::y, and mesh::Vector3::z.
Referenced by FemaxxDriver::run().
Here is the call graph for this function:
| void h5_write_eigenmesh | ( | const std::string & | file_name, | |
| const Epetra_Comm & | comm, | |||
| mesh::TetMesh * | tetmesh | |||
| ) |
Write the mesh used for the eigenvalue calculations into HDF5 file. The function must be called by alle processes
standard loop indices
store result code
global number of tetrahedra, distributed over processes
number of tetrahedra local to this process
store global point id
store tetrahedral vertex indices plus process id and number of processes engaged
Retrieve access to paralleltetmesh class instance
Manage HDF5 file operations
Open file collectively and release property list identifier
store the rank of this prcocess within this MPI communicator
store the size of an MPI communicator, i.e. the number of processes engaged
__DEBUG__VERBOSE__
Set up file access property list with parallel I/O access
Create a group named "/eigenmesh" in the file.
Create the dataspace for the dataset, including dataset dimensions and 2 additional integers, which are used to signal (i) the rank of the process which wrote a specific tetrahedron and the total number of processes engaged in the write operation; this information can later be used to signal tetrahedral mesh distribution over the processes by corresponding visualization.
Retrieve local number of tetrahedra
Create the dataset with default properties and close filespace; nota bene, if a new dataset shall be created within a group, then use the group's id when calling H5Dcreate!
Each process defines a dataset in memory and writes it to the hyperslab in the file
Store all columsn
Retrieve information to calculate offset of specific process within mpi communicator group
Array has mpi_size elements, stores number of tets over all processes
Compute offset for specific process mpi_rank
__DEBUG__VERBOSE__
Select the hyperslab within the file
Fill data buffer to be stored into the hyperslab; store tetrahedral vertex id's into a 2 dimensional array
We store the indices of the tetrahedron's vertices and addtionally, the rank of the MPI process writing this specific tetrahedron and also the number of processes engaged in this write up; therefore, the format is as follows:
id0 id1 id2 id3 rank size id0 id1 id2 id3 rank size id0 id1 id2 id3 rank size ... ... ... id0 id1 id2 id3 rank size id0 id1 id2 id3 rank size
Loop over tetrahedra and store them into HDF5 file
Increase index w.r.t tetrahedra
Create property list for collective dataset write
__DEBUG__VERBOSE__
__DEBUG__VERBOSE__
Free dynamically allocated data
Close and release resources
Definition at line 455 of file h5_tools.cpp.
References DIM2D, mesh::TetMesh::get_nof_tets(), mesh::ParallelTetMesh::get_num_global_tets(), H5_GROUP_FEMAX_EIGENMESH(), H5_NAME_FEMAX_TETVID(), NCORNERTET, mesh::ParallelTetMesh::point_gid(), mesh::TetMesh::tet_begin(), and mesh::TetMesh::tet_end().
Referenced by FemaxxDriver::run().
Here is the call graph for this function:
| void h5_write_eigenmodes | ( | const std::string & | file_name, | |
| const Epetra_MultiVector & | Q, | |||
| const Epetra_SerialDenseVector & | lambda | |||
| ) |
Read and write eigenmodes to HDF5 file.
This function must be called collectively.
Definition at line 297 of file h5_tools.cpp.
Referenced by FemaxxDriver::calculate_eigenfields().
| void h5_write_eigenpoint | ( | const std::string & | file_name, | |
| const Epetra_Comm & | comm, | |||
| mesh::TetMesh * | tetmesh | |||
| ) |
Write the tetrahedral point coordinates in parallel mode, ensuring unique file access and semantics.
Write the tetrahedral point coordinates into the HDF File; the function must be called collectively; to obtain a correct data layout, so that the tetrahedra defined previously are consistent with the order of the vertices stored, we use the hyperslab concept provided by HDF5.
The algorithm operates like this::
(i) loop over all poins that are accessible to this local process
(ii) find out if they belong to the respective process and mark them appropriately
(iii) write them into HDF5 file using the hyperslab approach
standard loop indices
store result code
global number of tetrahedra, distributed over processes
number of tetrahedra local to this process
store tetrahedral vertex indices plus process id and number of processes engaged
Retrieve access to paralleltetmesh class instance
Manage HDF5 file operations
Open file collectively and release property list identifier
store the rank of this prcocess within this MPI communicator
store the size of an MPI communicator, i.e. the number of processes engaged
Set up file access property list with parallel I/O access
Open the group named "/eigenmesh" in the file, if it exists; it not, then create it.
retrieve global number of points
Global number of points, i.e. sum of all number of points owned by the processes
Create the dataset with default properties and close filespace; nota bene, if a new dataset shall be created within a group, then use the group's id when calling H5Dcreate!
Each process defines a dataset in memory and writes it to the hyperslab in the file
retrieve global number of points
Number of points owned by this local process
Loop over points accesssible to the local process and find if the local process owns it
Retrieve number of points owned by process and create an array to store the coordinates
Loop over all points accessible to the local process
Create a complex hyperslab
Select the hyperslab within the file
Initialize data buffer
Increase number of points owned by process
increase point counter
__DEBUG__VERBOSE__
Create property list for collective dataset write
Free dynamically allocated data, if any
Close and release resources
Definition at line 638 of file h5_tools.cpp.
References DIM2D, mesh::TetMesh::get_nof_points(), mesh::ParallelTetMesh::get_num_global_points(), H5_GROUP_FEMAX_EIGENMESH(), H5_NAME_FEMAX_POINTS(), NCORNERTET, NREALCOORD3D, mesh::TetMesh::point_begin(), mesh::TetMesh::point_end(), and mesh::ParallelTetMesh::point_gid().
Referenced by FemaxxDriver::run().
Here is the call graph for this function:
| void h5_write_eigenvalue | ( | const std::string & | file_name, | |
| const Epetra_Comm & | comm, | |||
| mesh::TetMesh * | tetmesh, | |||
| NedelecMesh & | nedelecmesh, | |||
| const Epetra_MultiVector & | Q, | |||
| double * | lambda | |||
| ) |
Write the eigenvalues stored in 'lambda' into the HDF5 file.
Typically, the number of eigenvalues is much smaller than the number of elements present in an eigenvector, e.g. 5 or so, therefore, we just use the rank 0 process to write this tiny array into the HDF5 file; its number of rows can later be used to denote the number of eigenmodes present in the output HDF5 file by any postprocessing program. The eigenfrequency is calculate from the eigenvalue through = {} {c_{0}}{2 }.
store the rank of this prcocess within this MPI communicator
store the size of an MPI communicator, i.e. the number of processes engaged
Set up file access property list with parallel I/O access
Create the group named H5_GROUP_FEMAX_EIGENMODES in the file, if it exists; it not, then create it.
Create property list for collective dataset write
Definition at line 1482 of file h5_tools.cpp.
References DIM2D, h5_compute_eigenquality(), H5_GROUP_FEMAX_EIGENMODES(), H5_NAME_FEMAX_EIGENVALUE(), MPIROOTPROCESS, PI, and SPEED_OF_LIGHT_VACUUM.
Referenced by FemaxxDriver::run().
Here is the call graph for this function:
| void h5_write_param_list | ( | const std::string & | file_name, | |
| const Teuchos::ParameterList & | params | |||
| ) |
Write parameter list to HDF5 file.
Definition at line 202 of file h5_tools.cpp.
Referenced by FemaxxDriver::calculate_eigenfields().
 1.4.7
1.4.7